Overview¶
What is OpenFMO?¶
Open-architecture Implementation of Fragment Molecular Orbital Method for Peta-scale Computing (OpenFMO) [TMO+07][IMH+13] is an open-architecture program targeting the effective FMO calculations on massive parallel computers, now including post-Peta Scale systems.
Fragment molecular orbital (FMO) method [KIA+99][NKS+02][FK04] is a representative method to solve the electronic structures of large bio-molecules including protein, DNA, sugar chain, and so on.
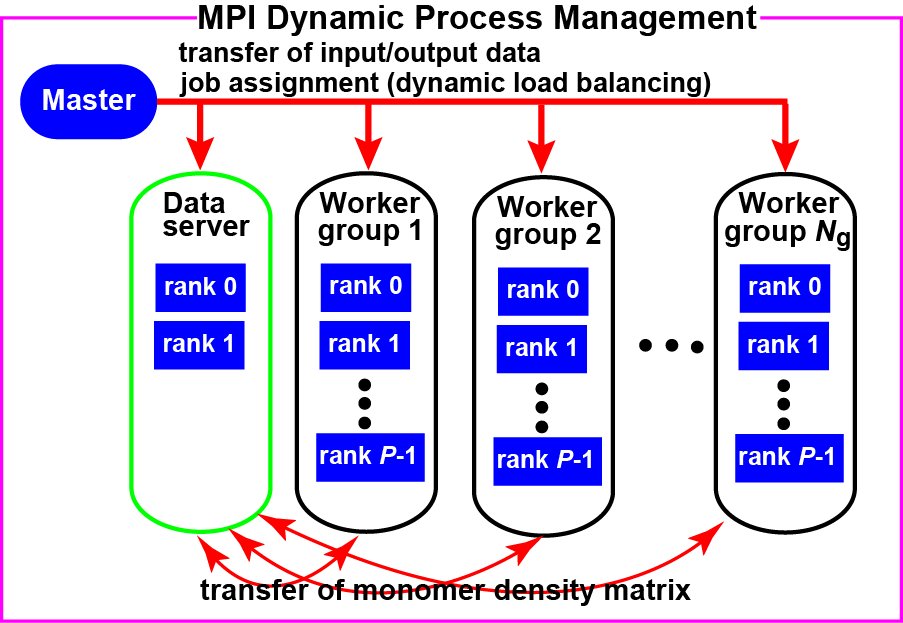
OpenFMO was written from scratch by Inadomi and co-workers [TMO+07][IMH+13] in C-language (ca. 54,000 lines) with OpneMP and MPI hybrid programming model. Figure 1 illustrates the MPI dynamic process management used for OpenFMO program on the basis of the master-worker execution model involving master process, worker groups, and data server.
In addition to FMO calculations, the users can do conventional restricted Hartree-Fock (RHF) calculations using the “skeleton-RHF” code of OpenFMO, which is also OpenMP and MPI hybrid program.
In this released version OpenFMO 1.0, GPU-accelerated FMO-RHF and RHF calculations can be performed on GPU cluster. [UHS+15b][UHS+15a][UHS+]
Capabilities and Limitations¶
- Single-point Ground-state Energy Calculation
- RHF with skeleton-RHF Code
- FMO2-RHF (FMO2: FMO Method with Two-body Correction)
- Minimum and Double-zeta Gaussian Basis Functions; Up to Third-row Atoms (namely, H - Ar)
- STO-3G, 6-31G, 6-31G(d), 6-31G(d,p)
- MPI + OpenMP Parallelization for RHF and FMO2-RHF
- GPU-accelerated RHF and FMO-RHF with Fermi or Kepler microarchitecture supporting double-precision floating-point operations
Citing OpenFMO¶
OpenFMO
Please cite the following references [TMO+07][IMH+13] for any publication including the scientific works obtained from OpenFMO application.
OpenFMO with Falanx Middleware
In addition, please cite the following reference [TIN+] for any publication including the scientific works obtained from OpenFMO application with Falanx middleware .
GPU-accelerated RHF
Please cite the following references [UHS+15b][UHS+] when publishing the results obtained from the GPU-accelerated skeleton RHF code in OpenFMO.
GPU-accelerated FMO-RHF
The following reference [UHS+15a][UHS+] should be cited when publishing the results utilizing the GPU-accelerated FMO-RHF code in OpenFMO.
License¶
OpenFMO:
Copyright (c) 2007 Yuichi Inadomi, Toshiya Takami, Hiroaki Honda, Jun Maki, and Mutsumi Aoyagi
Released under the MIT license
http://opensource.org/licenses/mit-license.php
The copy of the license is also included in the distribution as “LICENSE_OpenFMO”.
GPGPU parts:
Copyright (c) 2013 Hiroaki Umeda, Yuichi Inadomi, Toshihiro Hanawa, Mitsuo Shoji, Taisuke Boku, and Yasuteru Shigeta
Released under the MIT license
http://opensource.org/licenses/mit-license.php
The copy of the license is also included in the distribution as “LICENSE_GPGPU_parts”.
Falanx parts:
The Falanx middleware is copyrighted by National Institute Advanced Industrial Science and Technology (AIST), and is licensed under the Apache License, Version 2.0.
You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
which is also included in the distribution as “LICENSE_Falanx”.
Released Version 1.0:
Copyright (c) 2018 Hiroaki Umeda, Yuichi Inadomi, Hirotaka Kitoh-Nishioka, and Yasuteru Shigeta
Released under the MIT license
http://opensource.org/licenses/mit-license.php
The copy of the license is also included in the distribution as “LICENSE_Released_Version_1.0”.
Contact Information¶
If you have a question about OpenFMO or find any bugs, please contact Hirotaka Kitoh-Nishioka at Biological Function and Information Group of Professor Shigeta in Center for Computational Sciences, University of Tsukuba.
Acknowledgments¶
JST-CREST: “Development of System Software Technologies for post-Peta Scale High Performance Computing”
| [FK04] | Dmitri G. Fedorov and Kazuo Kitaura. The importance of three-body terms in the fragment molecular orbital method. J. Chem. Phys., 120(15):6832–6840, 2004. URL: http://aip.scitation.org/doi/abs/10.1063/1.1687334, arXiv:http://aip.scitation.org/doi/pdf/10.1063/1.1687334, doi:10.1063/1.1687334. |
| [IMH+13] | (1, 2, 3) Yuichi Inadomi, Jun Maki, Hiroaki Honda, Toshiya Takami, Taizo Kobayashi, Mutsumi Aoyagi, and Kasuo Minami. Performance tuning of parallel fragment molecular orbital program (openfmo) for effective execution on k-computer. J. Comput. Chem., Jpn., 12(2):145–155, 2013. doi:10.2477/jccj.2012-0031. |
| [KIA+99] | Kazuo Kitaura, Eiji Ikeo, Toshio Asada, Tatsuya Nakano, and Masami Uebayasi. Fragment molecular orbital method: an approximate computational method for large molecules. Chem. Phys. Lett., 313(3–4):701 – 706, 1999. URL: http://www.sciencedirect.com/science/article/pii/S000926149900874X, doi:http://dx.doi.org/10.1016/S0009-2614(99)00874-X. |
| [NKS+02] | Tatsuya Nakano, Tsuguchika Kaminuma, Toshiyuki Sato, Kaori Fukuzawa, Yutaka Akiyama, Masami Uebayasi, and Kazuo Kitaura. Fragment molecular orbital method: use of approximate electrostatic potential. Chem. Phys. Lett., 351(5–6):475 – 480, 2002. URL: http://www.sciencedirect.com/science/article/pii/S0009261401014166, doi:http://dx.doi.org/10.1016/S0009-2614(01)01416-6. |
| [TMO+07] | (1, 2, 3) Toshiya Takami, Jun Maki, Jun-ichi Ooba, Yuichi Inadomi, Hiroaki Honda, Taizo Kobayashi, Rie Nogita, and Mutsumi Aoyagi. Open-architecture implementation of fragment molecular orbital method for peta-scale computing. CoRR, 2007. URL: http://arxiv.org/abs/cs/0701075, arXiv:cs/0701075. |
| [TIN+] | Atsuko Takefusa, Tsutomu Ikegami, Hidemoto Nakada, Ryousei Takano, Takayuki Tozawa, and Yoshio Tanaka. Scalable and highly available fault resilient programming middleware for exascale computing. The International Conference for High Performance Computing, Networking, Storage and Analysis: SC14. URL: http://sc14.supercomputing.org/sites/all/themes/sc14/files/archive/tech_poster/poster_files/post217s2-file2.pdf. |
| [UHS+] | (1, 2, 3) Hiroaki Umeda, Toshihiro Hanawa, Mitsuo Shoji, Taisuke Boku, and Yasuteru Shigeta. Large-scale mo calculation with gpu-accelerated fmo program. The International Conference for High Performance Computing, Networking, Storage and Analysis: SC15. URL: http://sc15.supercomputing.org/sites/all/themes/SC15images/tech_poster/tech_poster_pages/post198.html. |
| [UHS+15a] | (1, 2) Hiroaki Umeda, Toshihiro Hanawa, Mitsuo Shoji, Taisuke Boku, and Yasuteru Shigeta. Gpu-accelerated fmo calculation with openfmo: four-center inter-fragment coulomb interaction. J. Comput. Chem., Jpn., 14(3):69–70, 2015. doi:10.2477/jccj.2015-0041. |
| [UHS+15b] | (1, 2) Hiroaki Umeda, Toshihiro Hanawa, Mitsuo Shoji, Taisuke Boku, and Yasuteru Shigeta. Performance benchmark of fmo calculation with gpu-accelerated fock matrix preparation routine. J. Comput. Chem., Jpn., 13(6):323–324, 2015. doi:10.2477/jccj.2014-0053. |